
Salvador Dura-Bernal, PhD
Associate Professor
Physiology and Pharmacology
In the Dura-Bernal lab, we use computational simulations and mathematical models to better understand neural brain function and disorders, and help develop novel treatments. We build and simulate large-scale biophysically detailed models of the brain neural circuits linking multiple scales: molecules, cells, networks and systems. We run these simulations on supercomputers to reproduce experimental data and make predictions. We develop software tools to facilitate building these models, including NetPyNE, which has been used in over 40 labs (netpyne.org). We also use machine learning to optimize, analyze and interpret neuroscience data, and develop brain-machine interfaces based on these biologically-detailed circuit models.
Multiscale Modeling of Cortical Circuits
Neural coding mechanisms from molecules to behavior: We integrate experimental data into detailed models to study the interactions across scales (molecular, cellular, circuit and behavior) and their role in brain function and disease. For example, how the interactions between long-range thalamocortical inputs and noradrenergic modulation of HCN current regulates M1 dynamics associated with behavior. We use models to identify and characterize neural coding mechanisms within and across scales, including synchronous firing of neuronal ensembles, phase-amplitude oscillatory coupling, dendritic and network resonance, neuromodulatory interactions (e.g. noradrenaline and dopamine), neuronal avalanches, and information flow.
Models of thalamocortical circuits: We developed a model of mouse primary motor cortex circuits containing over 10,000 biophysically and morphologically realistic neurons and 30 million synapses, with neuronal density, distribution and synaptic connectivity patterns derived from experimental data; and validated different cell type-specific dynamics associated with behavior against in vivo data. In collaboration with the Nathan Kline Institute, we built a biologically-detailed model of macaque auditory thalamocortical circuits, receiving auditory input from a model of cochlea and inferior colliculus, to investigate the mechanistic origins of spatiotemporal oscillatory patterns observed in vivo. Preliminary simulation results matched in vivo macaque firing rates and LFP/CSD oscillations.
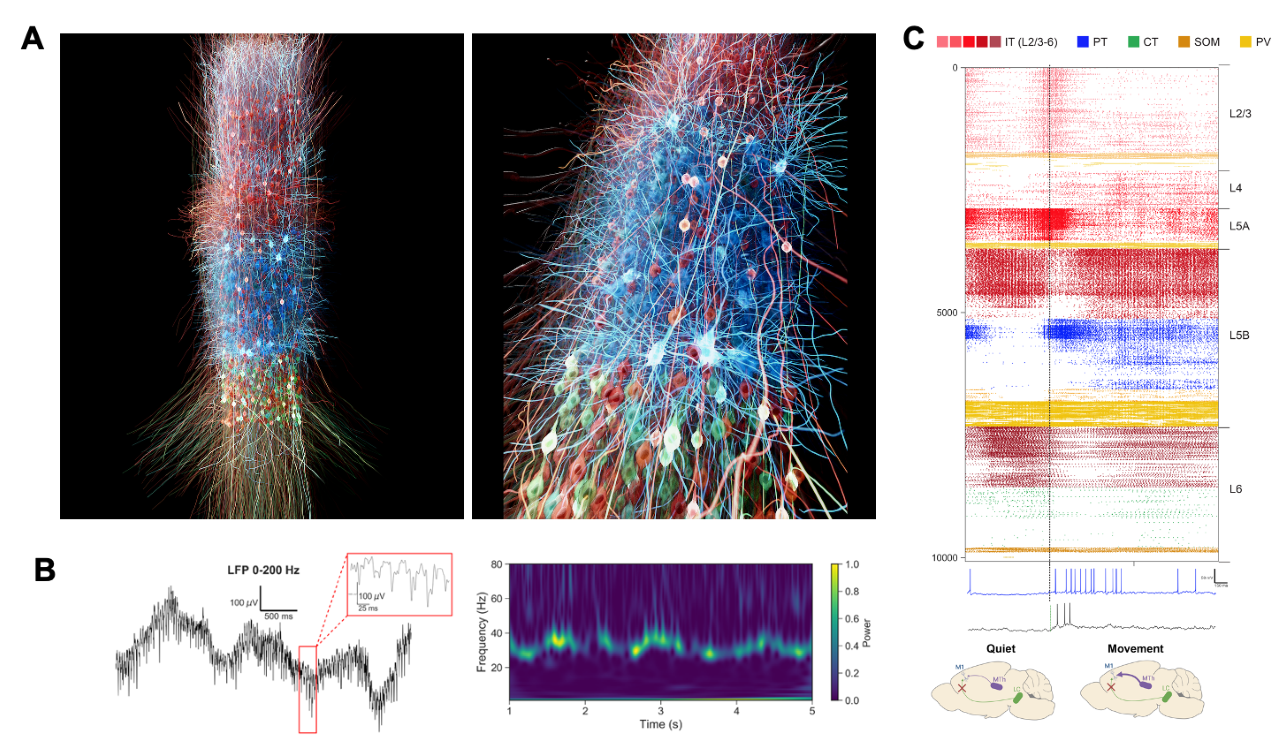
Fig 1. Detailed model of mouse motor cortex (M1) circuits. A) 3D visualization of M1 network, showing location and morphologies of excitatory IT (red), PT (blue) and CT (green) cells, and snapshot of simulated activity with spiking neurons in brighter color (visualization by nicolasantille.com). B) Simulated M1 local field potential (LFP) raw signal and spectrogram showing cross-frequency coupling of delta and gamma oscillations. C) M1 cell-type and layer-specific firing dynamics associated with the quiet and movement behavioral states, during the noradrenaline (NA) block condition (decreased ih current in PT cells).
Software Tools for Modeling
Tool for multiscale modeling of brain circuits: NetPyNE is an open-source Python package to facilitate the development, simulation, parallelization, analysis, and optimization of biological neuronal networks using the NEURON simulator. It includes a state-of-the-art graphical user interface developed in collaboration with MetaCell. More info at www.netpyne.org
Collaborations with other tools and resources: We collaborate with multiple other groups in the development of tools, resources and standards to advance computational neuroscience, including OpenSourceBrain, NeuroML, SONATA, Geppetto, Human Neocortical Neurosolver, Neuroscience Gateway, E-BRAINS, coreNEURON, SciDash and The Virtual Brain.
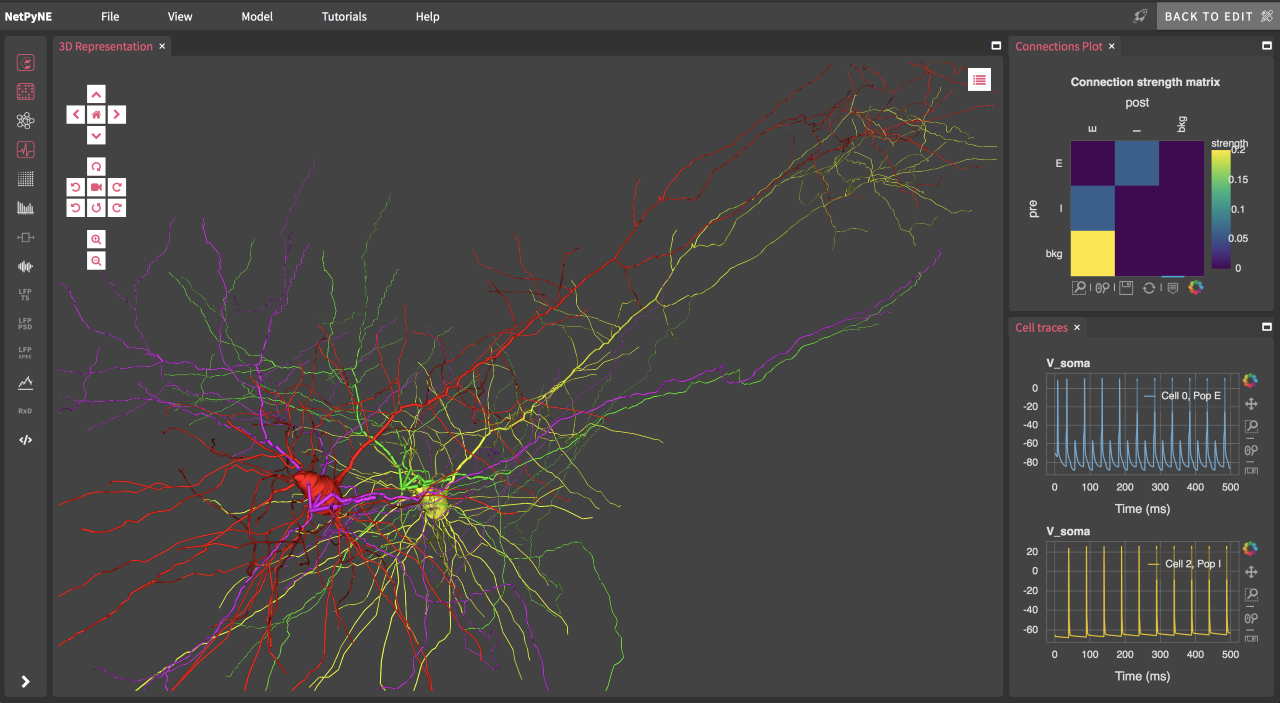
Fig 2. Graphical user interface of NetPyNE tool used to develop data-driven multiscale models of brain circuits (www.netpyne.org)
Machine Learning for Neuroscience
Optimization and analysis of detailed cortical models: We use machine learning to analyze the neural model structure and simulation output (e.g. clustering of neuronal avalanches) and to automatically optimize the model parameters to reproduce experimental data (e.g. via genetic/evolutionary algorithms).
Deep learning models of cortical systems: We develop deep learning models, including convolutional networks and Bayesian networks with belief propagation, to better understand the function of circuits underlying sensory perception. Models are based on anatomical and physiological experimental data and reproduce features of visual and auditory cortex dynamics.
Biomimetic Neuroprosthetics
We develop biomimetic brain-machine interfaces based on realistic cortical spiking models that can interact directly with the biological neurons and with a robotic/virtual arm in real time. The system could be used to decode the patient's brain signals through a more biological approach, exploiting plasticity and adaption, and could potentially replace damaged brain regions with in silico counterparts. We also use our multiscale brain models to simulate the effects of neurostimulation (e.g. electrical or optogenetic) on neural circuits, and help predict the stimulation patterns required to alter network dynamics and restore healthy function for different brain lesions or disorders.
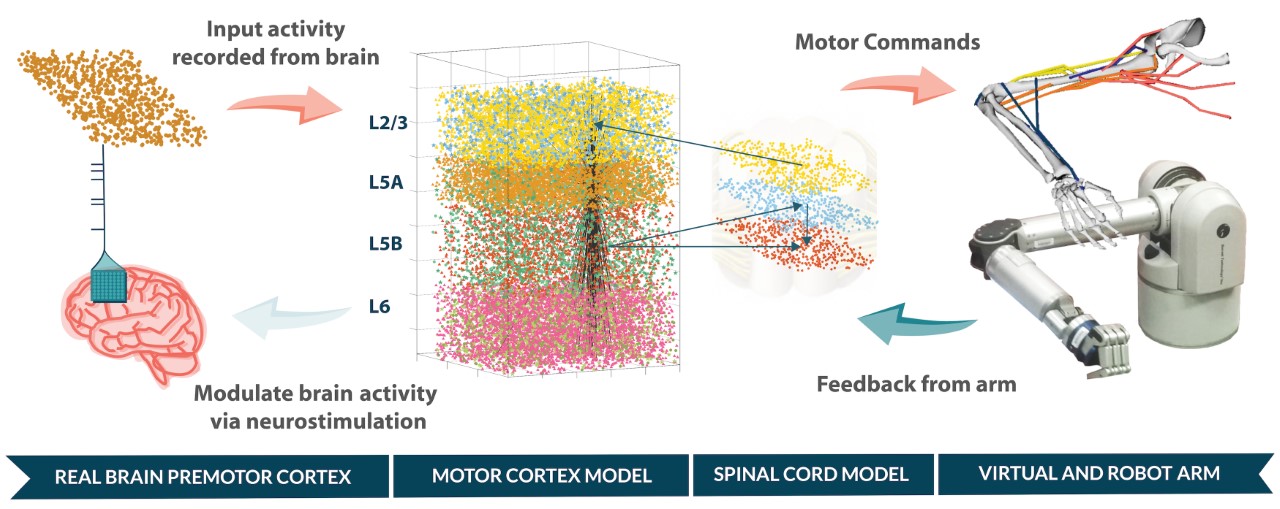
Fig 3. Design for neuroprosthetic system to replace damaged cortical region.A motor cortex model was interfaced with brain signals and with a robot and virtual arm in real-time. The network included biological plasticity and reinforcement learning. The system was trained to control the arm using evolutionary algorithms running on supercomputers.
For further information, see Dura-Bernal Lab website.
Personnel
James Chen, Ph.D., Postdoctoral Fellow
Roman Baravalle, Ph.D., Postdoctoral Fellow
Nikita Novikov, Ph.D., Postdoctoral Fellow
Joao V Moreira, M.Sc., Graduate Student
Scott McElroy B.S., Graduate Student
Molly Leitner B.S., Graduate Student
Service Functions
Interim Co-Director of the Global Center for AI in Mental Health
Elected to Board of Directors of the Organization for Computational Neurosciences.
Elected Member of the NeuroML Editorial Board.
Reviewer for multiple NIH study sections (BRAIN Initiative and Sensorimotor Integration).
Editor and reviewer for multiple scientific journals.
- Dura-Bernal, S., Kan, L., Neymotin, S. A., Francis, J.T ., Principe, J. C., and Lytton, W. W. (2016). Restoring behavior via inverse controller in a lesioned cortical spiking model driving a virtual arm. Frontiers in Neuroscience (Neuroprosthetics) 10, 28.
- Dura-Bernal, S., Neymotin, S. A., Kerr, C. C., Sivagnanam, S., Majumdar, A., Francis, J. T., and Lytton, W. W. (2017) Evolutionary algorithm optimization of biological learning parameters in a biomimetic neuroprosthesis. IBM J. Res. Dev., 61:2/3.
- Alber, M., Buganza Tepole, A., Cannon, W. R., De, S., Dura-Bernal, S., Garikipati, K., Karniadakis, G., Lytton, W. W., Perdikaris, P., Petzold, L., and Kuhl, E. (2019). Integrating machine learning and multiscale modeling-perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. NPJ Digit Med 2, 115.
- Dura-Bernal, S., Suter, B.A., Gleeson, P., Cantarelli, M., Quintana, A., Rodriguez, F., Kedziora, D. J., Chadderdon, G. L., Kerr, C. C., Neymotin, S. A., McDougal, R. A., Hines, M., Shepherd, G. M. G., and Lytton W.W. (2019). NetPyNE, a tool for data-driven multiscale modeling of brain circuits. eLife 8:e44494.
- Romaro, C., Najman, F., Lytton, W. W., Roque, A. C., and Dura-Bernal, S. (2021). NetPyNE implementation and rescaling of the Potjans-Diesmann cortical microcircuit model. Neural Computation 33, 1993–2032.
- Borges, F., Moreira, J., Takarabe, L. M., Lytton, W. W., and Dura-Bernal, S. (2022). Large-scale biophysically detailed model of somatosensory thalamocortical circuits in NetPyNE. Frontiers in Neuroinformatics doi: 10.3389/fninf.2022.884245.
- Dura-Bernal, S., Neymotin, S. A., Suter, B. A., Dacre, J., Moreira, J. V. S., Urdapilleta, E., Schiemann, J., Duguid, I., Shepherd, G. M. G., and Lytton, W.W. (2023). Multiscale model of primary motor cortex circuits predicts in vivo cell-type-specific, behavioral state-dependent dynamics. Cell Reports 42, 112574.
- Dura-Bernal, S., Griffith, E. Y., Barczak, A., O’Connell, M. N., McGinnis, T., Moreira, J. V. S., Schroeder, C. E., Lytton, W. W., Lakatos, P., and Neymotin, S. A. (2023). Data-driven multiscale model of macaque auditory thalamocortical circuits reproduces in vivo dynamics. Cell Reports 42, 10.1016/j.celrep.2023.113378.
- Varela, .C, Moreira, J. V. S., Kocaoglu, B., Dura-Bernal, S., and Ahmad, S. (2024). A mechanism for deviance detection and contextual routing in the thalamus: a review and theoretical proposal. Frontiers in Neuroscience 18. doi: 10.3389/fnins.2024.1359180.
- Sinha, A., Gleeson, P., Marin, B., Dura-Bernal, S., Panagiotou, S., Crook, S., Cantarelli, M., Cannon, R. C., Davison, A. P., Gurnani, H., and Silver, R. A. (2024). The NeuroML ecosystem for standardized multi-scale modeling in neuroscience. eLife 13:RP95135.
List of Publications (Pub Med) https://www.ncbi.nlm.nih.gov/myncbi/16IxrzkzJBFQm/bibliography/public/