
Miriam H. Feuerman, PhD
Associate Professor
Cell Biology
The normal adult liver has the unique ability to regenerate after poisoning, surgical resection or viral damage. However, repeated growth cycles as observed in chronic Hepatitis B Virus (HBV) or Hepatitis C Virus infection, can result in the failure of the normal controls on liver regeneration, leading to liver cancer. The alpha-fetoprotein (AFP) gene, an oncofetal antigen, is expressed in the developing fetus and under conditions of liver regeneration and tumorigenesis in the adult. The level of AFP gene expression during liver regeneration in mice is regulated by a genetically unlinked autosomal locus, Afr2. Inbred C57BL/6 mice express 8 to 10 fold less AFP during liver regeneration than wild type C3H/He mice. Lower AFP expression in regenerating liver reflects other regulatory processes, as C57BL/6 mice are less susceptible to liver carcinogenesis than C3H/He mice. The mechanisms that regulate growth may also regulate AFP gene expression making it an ideal surrogate marker to monitor the condition of the liver.
Analysis of expression of a panel of transgenic mice allowed us to identify the region of the AFP gene locus required for Afr2 regulation and Afr2 Response Element or ARE. Subsequent work in hepatoma cell lines, primary hepatocytes and in mice shows that this sequence does not function as a standard transcription factor recognition site in the same fashion as much of the AFP gene locus, although there are many sequences known to be recognized by transcription factors.
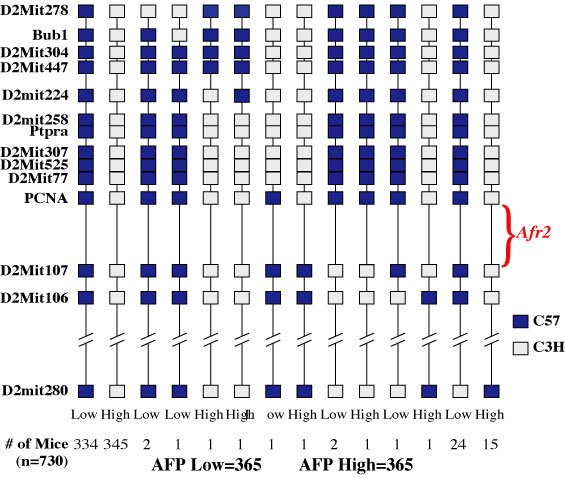 |
| Figure 1. Genetic mapping of the Afr2 gene locus. 730 mice were analyzed to determine Afr2 phenotype and genetic composition. The Afr2 gene locus must reside between PCNA and D2Mit107, an anonymous marker identified by the Mouse Genome Project. |
 |
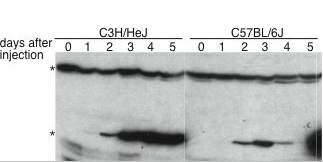 |
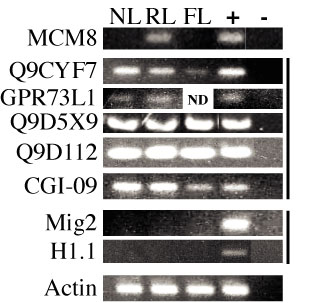
| Figure 2. Pattern of gene expression for Afr2 candidate genes. RT-PCR was used to determine the abundance of mRNA for each of the genes located within the Afr2 gene locus. | Figure 3. MCM8 expression over the course of liver regeneration. Mice were injected with CCl4 and sacrificed at the days indicated after injection. 100 mg of whole cell extracts were subject to western blot analysis with anti-MCM8 antiserum. |
- Pan, X., F.N. Hussain, J. Iqbal, M.H. Feuerman and M. M Hussain (2007). Inhibiting proteasomal degradation of microsomal triglyceride transfer protein prevents CCl4 induced steatosis. Journal of Biological Chemistry 282(23):17078-89
- Cui, R., T.T. Nguyen, J.H. Taube, S.A, Statton, M.H. Feuerman and M.C. Barton (2005) Family members p53 and p73 act together in chromatin modification and direct repression of AFP transcription. Journal of Biological Chemistry 280(47):39152-60
- Park, J. K. and M.H. Feuerman (2005). Afr2 regulation occurs cell-autonomously in vitro but is not conferred on episomal DNA in transient assays. DNA and Cell Biology 24(3): 189-198
- Jin, D.K., E.C. Anderson, E. Gilbert and M.H. Feuerman (2005). AFP gene expression acute diethylnitrosame intoxication is not Afr2 regulated. Cancer Letters 220 (2):211-220
- Feuerman, M.H. (2002) Alpha-fetoprotein. Encyclopedia of Molecular Medicine. Editor: Thomas E. Creighton, EMBL. Editorial Board: C. Thomas Caskey, Michael R. Hayden, Haig H. Kakazian, Jr., George Klein, Hugo W. Moser, Anthony J. Pawson, Stuart H. Orkin, Bernard Roizman, R.V. Thakker, Hugh Watkins. Publisher John Wiley and Sons, New York
- Jin, D.K., J. Vacher and M.H. Feuerman (1998). Alpha-fetoprotein Sequences Mediating Afr2 Regulation during Liver Regeneration. Proceedings of the National Academy of Science 95 (15): 8768-8772.
- Jin, D.K. and M.H. Feuerman (1998). Chromosomal location of Afr2, regulator of alpha-fetoprotein gene expression during liver regeneration. Mammalian Genome. 9:256-258.